By, Tessa E. Reid, Vanessa N. Kavamura, Maïder Abadie, Adriana Torres-Ballesteros, Mark Pawlett, Ian M. Clark, Jim Harris, Tim H. Mauchline.
The profound negative effect of inorganic chemical fertilizer application on rhizobacterial diversity has been well documented using 16S rRNA gene amplicon sequencing and predictive metagenomics. We aimed to measure the function and relative abundance of readily culturable putative plant growth-promoting rhizobacterial (PGPR) isolates from wheat root soil samples under contrasting inorganic fertilization regimes. We hypothesized that putative PGPR abundance will be reduced in fertilized relative to unfertilized samples. Triticum aestivum cv.
Cadenza seeds were sown in a nutrient depleted agricultural soil in pots treated with and without Osmocote® fertilizer containing nitrogen-phosphorous-potassium (NPK). Rhizosphere and rhizoplane samples were collected at flowering stage (10 weeks) and analyzed by culture-independent (CI) amplicon sequence variant (ASV) analysis of rhizobacterial DNA as well as culture-dependent (CD) techniques.
Rhizosphere and rhizoplane derived microbiota culture collections were tested for plant growth-promoting traits using functional bioassays. In general, fertilizer addition decreased the proportion of nutrient-solubilizing bacteria (nitrate, phosphate, potassium, iron, and zinc) isolated from rhizocompartments in wheat whereas salt tolerant bacteria were not affected.
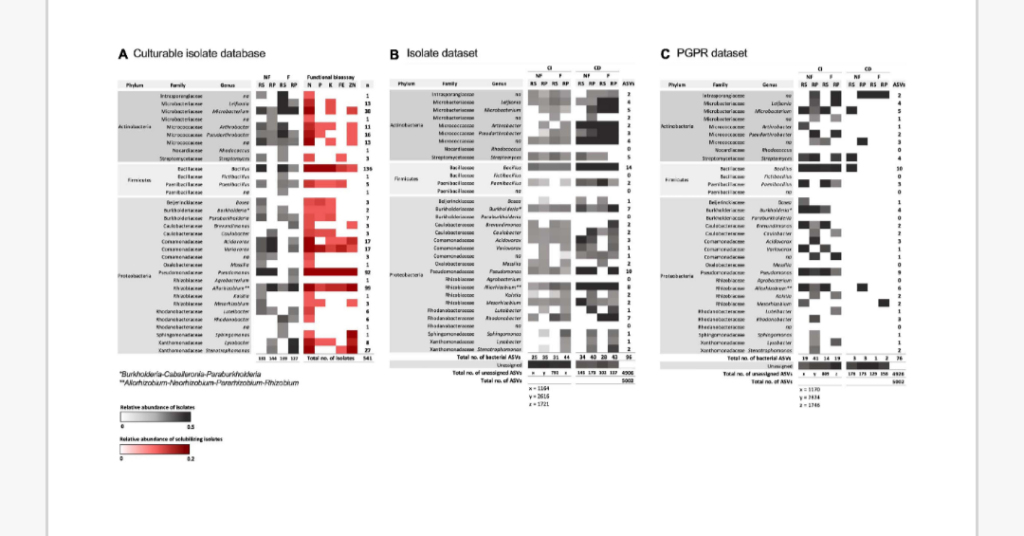
A “PGPR” database was created from isolate 16S rRNA gene sequences against which total amplified 16S rRNA soil DNA was searched, identifying 1.52% of total community ASVs as culturable PGPR isolates. Bioassays identified a higher proportion of PGPR in non-fertilized samples [rhizosphere (49%) and rhizoplane (91%)] compared to fertilized samples [rhizosphere (21%) and rhizoplane (19%)] which constituted approximately 1.95 and 1.25% in non-fertilized and fertilized total community DNA, respectively.
The analyses of 16S rRNA genes and deduced functional profiles provide an in-depth understanding of the responses of bacterial communities to fertilizer; our study suggests that rhizobacteria that potentially benefit plants by mobilizing insoluble nutrients in soil are reduced by chemical fertilizer addition. This knowledge will benefit the development of more targeted biofertilization strategies.
Introduction
Since anthropogenic plant domestication began ca. 19,000 years ago, edible plants, e.g., cereals have been extensively bred (Lev-Yadun et al., 2000; Tanno and Willcox, 2006).
However, current high-yielding dwarf crop varieties rely on unsustainable levels of inorganic nitrogen and phosphorous fertilizers, pesticides, and other chemical inputs which are environmentally harmful (Rees et al., 2013; Van Grinsven et al., 2013). Wheat is the third most cultivated cereal in the world; the Cambric Arenosol (FAO) predicts that by 2050 the global population will reach 9.73 billion meaning food production must be accordingly increased by 50% (FAO, 2017). By 2027, demand for wheat will increase to 833 million tons, which is 10% above the annual current production (OECD/FAO, 2018).
Soil microbial communities influence plant growth, health, and resource use efficiency, especially the subset that coexist and are selected to form the root microbiome (Berendson et al., 2012; Mendes et al., 2013; Schlaeppi and Bulgarelli, 2015; Mauchline and Malone, 2017). A gradient of intimacy between plant roots and microbes extends away from the root: plant influence over the microbial community increases nearer the root surface. Root surface microbes are said to inhabit the rhizoplane, and those in soil closely associated with the root, the rhizosphere (Hiltner, 1904; Zhang et al., 2017).
Rhizosphere and rhizoplane microorganisms can benefit crop plants in several ways including improved plant nutritional status and protection against biotic and abiotic stresses (Bloemberg and Lugtenberg, 2001; Turner et al., 2013; Choudhary et al., 2016; Ahkami et al., 2017), so are termed plant growth-promoting rhizobacteria (PGPR) (Kloepper and Schroth, 1978). Inoculating plants with PGPR can stimulate crop growth, forming the basis for the biofertilizer industry providing green alternatives to synthetic fertilizers and agrochemicals (Backer et al., 2018). Understanding the effect of agricultural practices, such as fertilization regime, on PGPR populations is essential to optimize microbiome function in the sustainable intensification of agriculture (Hartmann et al., 2015).
To date, microbial community studies have focused on taxonomic composition, but the functional potential of the microbiome may be more important to ensure key functions for holobiont fitness (Bai et al., 2015; Lemanceau et al., 2017).
With advances in next-generation sequencing technologies, the wheat microbiome has mostly been defined based on culture-independent (CI) methods; host genotype (Mahoney et al., 2017), fertilization regime (Kavamura et al., 2018; Chen et al., 2019), land management and seed load (Kavamura et al., 2019), irrigation (Mavrodi et al., 2018), growth stage (Chen et al., 2019), and dwarfing (Kavamura et al., 2020) all affect the rhizosphere community structure.
However, little has been done to link taxonomic structure of the wheat microbiome to its functional ability.
Using CI and culture-dependent (CD) methods, we studied the effect of chemical fertilizer on putative PGPR abundance in the commercial wheat variety, Cadenza, from a low input agricultural soil depleted in most nutrients, in which beneficial microorganisms are important to sustain crop production.
We hypothesized that the abundance of rhizobacteria with plant growth-promoting traits would be reduced for fertilized relative to unfertilized wheat due to differences in plant nutrient status. Addition of nitrogen-phosphorous-potassium (NPK) would mean that plants no longer need to interact with beneficial rhizobacteria to provide nutrients to sustain growth.
Our aim was to characterize culturable bacteria with plant growth-promoting traits; determine their abundance within culturable communities; and characterize CI and CD 16S rRNA gene DNA to assess the impact of the widely used NPK fertilizer on putative PGPR populations.
This was achieved by creating isolate libraries from each soil sample and subjecting them to a range of bioassays which test key traits in nutrient acquisition to establish the abundance of isolates with beneficial traits. 16S rRNA gene sequences from isolates were used to create a “PGPR” database against which total CI amplified 16S rRNA soil DNA was searched in order to determine the relative abundance of culturable PGPR within total community DNA.
Materials and Methods
Soil Collection, Experimental Setup, and Harvesting
We evaluated the rhizosphere and rhizoplane soil from wheat grown with and without NPK fertilizer. Soil was collected from Stackyard bare-fallow soil mine (LATLONG 52.000293N, -0.614308E), a well-draining sandy loam soil from the Rothamsted Research experimental farm at Woburn, Bedfordshire (United Kingdom).
The soil is a Cottenham series (Catt et al., 1980) classified as a FAO, chosen to reduce the legacy effect of prior cropping systems. Soil was sieved (2 mm mesh), mixed thoroughly, and stored at 4°C in polythene bags prior to use.
Triticum aestivum cv. Cadenza seeds were surface sterilized [70% ethanol, 10 min; 1.5% active chlorine, 1 h; 5 × rinse, sterile distilled water (SDW); overnight imbibition, sterile water, 4°C] before germination on filter paper. Seedlings were planted (1× seedling/pot; 9 × 9 × 10 cm pots, ∼500 g soil) with and without NPK granules [15% N, 9% P2O5, 11% K2O, 2% MgO with micro-nutrients (B, Cu, Fe, Mn, Mo, and Zn); Osmocote, United Kingdom] (∼2.5 g per pot). Four replicate pots were prepared for each treatment. Plants were grown in a glasshouse (20°C, 16 h/day light regime) and watered daily with tap water.
Pots were harvested at the start of flowering (Zadoks growth stage 61; 10 weeks post germination) (Zadoks et al., 1974), resulting in eight rhizosphere samples and eight root samples. Height (from soil surface to head of longest stem) was measured, then soil was gently tipped from the pot onto a fresh polythene bag. Loose soil was discarded and non-rhizospheric soil carefully removed. Roots were vigorously shaken in a bag to release tightly attached soil (i.e., rhizosphere) and mixed to homogenize.
The root system was excised, cut vertically in half and placed in sterile 10 ml vials for subsequent rhizoplane work. One half was frozen (−20°C) for soil DNA extraction and the other stored (4°C) for culture work. Around 5 g of rhizosphere soil and 1 g of root was collected per plant. The remainder of the plant was dried (80°C, 24 h) and dry foliar plant biomass measured.
Isolation of Bacteria
To obtain a library of rhizospheric bacteria, 1 g of each rhizosphere soil sample was diluted 10-fold (SDW) and shaken vigorously for 10 min using a shaker.
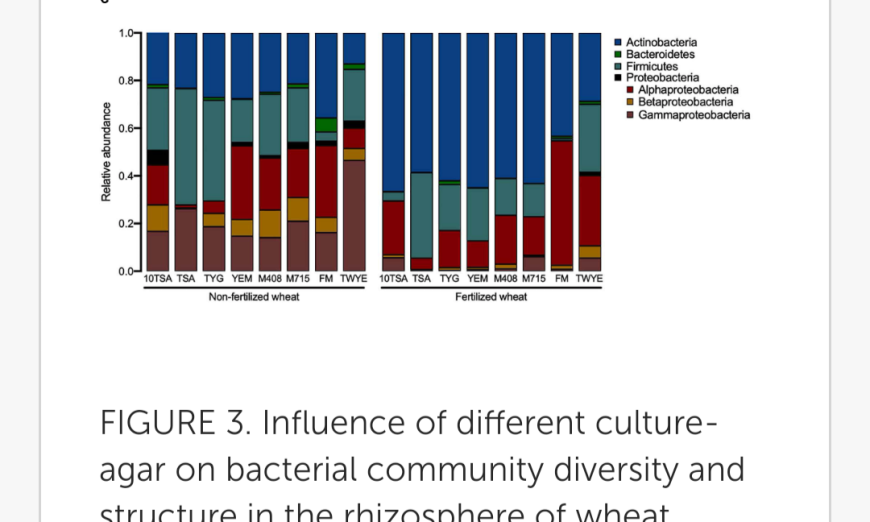
To increase the diversity and number of culturable isolates returned in this study, rhizosphere samples were plated onto both 10TSA and an additional seven agar types (Supplementary Table 1; Bai et al., 2015). Soil suspensions were serially diluted and 100 μl of final dilution spread on agar (six replicates per agar type). Different dilutions were plated depending on agar type (Supplementary Table 2) and incubated (25°C) for 4 days. Individual colonies were picked and inoculated in 500 μl tryptone soya broth (TSB) (Oxoid, Basingstoke, United Kingdom) (1/10th concentration) in deep well 96-well plates (Supplementary Figure 1) and incubated (25°C, 2 days).
For rhizoplane-colonizing bacteria, root samples were weighed and 900 μl SDW was added for every 0.1 g root. Samples were shaken vigorously for 10 min using a shaker, serially diluted, plated onto 10TSA and incubated (25°C, 4–6 days). Individual colonies were picked and inoculated in 500 μl TSB (1/10th concentration) (96-well plates, 25°C, 2 days) prior to functional analysis.
In addition, once colonies had been hand-picked from agar plates the remaining microbial biomass was resuspended in 5 ml SDW using a sterile spreader and transferred to a 50 ml centrifuge tube. Suspensions from technical replicates of the same agar (Supplementary Figure 2) were combined and each sample vortexed (10 min, high speed) to ensure homogeneity. Sub-samples of each were stored (2 ml Eppendorf tube, −20°C) for genomic DNA extraction.
ORIGINAL RESEARCH Article.
Front. Microbiol., 11 March 2021.
- Sustainable Agriculture Sciences, Rothamsted Research, Harpenden, United Kingdom.
- Cranfield Soil and Agrifood Institute, Cranfield University, Cranfield, United Kingdom. Data Availability Statement
The raw sequences analyzed for this study can be found in the NCBI Sequence Read Archive (SRA), accession PRJNA625513 and in NCBI Genbank, accessions MT354024 – MT354564.
Author Contributions.
TR, TM, IC, and VK designed the experiments. TR, VK, and MA performed the experiments and collected the data. TR analyzed the data with input from AT-B, VK, and MA for bioinformatic analyses. TR wrote the manuscript. All co-authors edited and commented on the manuscript.
Acknowledgments.
The authors would like to thank Ahmed Abdullah and Jess Evans for input with statistical analysis.
Citation: Reid TE, Kavamura VN, Abadie M, Torres-Ballesteros A, Pawlett M, Clark IM, Harris J and Mauchline TH (2021) Inorganic Chemical Fertilizer Application to Wheat Reduces the Abundance of Putative Plant Growth-Promoting Rhizobacteria. Front. Microbiol. 12:642587. doi: 10.3389/fmicb.2021.642587
Received: 16 December 2020; Accepted: 15 February 2021;
Published: 11 March 2021.
Edited by:
Aleksa Obradović, University of Belgrade, Serbia
Reviewed by,
Abhishek Walia, Chaudhary Sarwan Kumar, Himachal Pradesh Krishi Vishvavidyalaya, India.
Aymé Spor, INRA UMR 1347 Agroécologie, France.
Copyright © 2021 Reid, Kavamura, Abadie, Torres-Ballesteros, Pawlett, Clark, Harris and Mauchline.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Courtesy and thanks to Frontiers, the original publishers.
.